AutoLFADS on NeuroCAAS
Analysis Description
Latent factor analysis via dynamical systems (LFADS) is a variational sequential autoencoder that achieves state-of-the-art performance in denoising high-dimensional neural spiking activity for downstream applications in science and engineering. Recently introduced variants have continued to demonstrate the applicability of the architecture to a wide variety of problems in neuroscience.
Since the development of the original implementation of LFADS, new technologies have emerged that use dynamic computation graphs, minimize boilerplate code, compose model configuration files, and simplify large-scale training. This implementation of LFADS, lfads-torch, builds on these modern Python libraries and is designed to be easier to understand, configure, and extend. Note that the repo is currently private pending some internal benchmarking, but we plan to make it public soon.
Achieving state-of-the-art performance with deep neural population dynamics models requires extensive hyperparameter tuning for each dataset. AutoLFADS is a model-tuning framework that automatically produces high-performing LFADS models on data from a variety of brain areas and tasks, without behavioral or task information. The AutoLFADS framework uses coordinated dropout to prevent identity overfitting and population-based training to efficiently tune hyperparameters over the course of a single training run.
Preparing Data and Configuration Files for Analysis
NeuroCAAS runs the neurocaas branch of lfads-torch, which will be publicly available at the link above. At a high level, the pipeline takes (1) an input data file in the HDF5 format along with (2) a configuration YAML file which specifies model architecture and training hyperparameters. At the end of training, the pipeline will return a .zip file which contains training logs, model outputs, and the best performing model checkpoint.
Data File
At a minimum, the HDF5 data file must have the following keys, in the n_trials x n_timesteps x n_channels format:
train_encod_data: Data to be used as input when training the model.train_recon_data: Data to be used as a reconstruction target when training the model.valid_encod_data: Data to be used as input when validating the model.valid_recon_data: Data to be used as a reconstruction target when validating the model.
Configuration File
Basic Configuration
Starting from the demo config file, you'll need to edit the following fields to get the code to run:
encod_data_dim: Then_channelsdimension ofencod_datafrom your data file.encod_seq_len: Then_timestepsdimension ofencod_datafrom your data file.recon_seq_len: Then_timestepsdimension ofrecon_datafrom your data file.readout.modules.0.out_features: Then_channelsdimension ofrecon_datafrom your data file.datamodule.batch_size: The batch size to use for training and inference. Should mainly be used to manage memory usage. Two models per GPU is typical.datamodule.batch_keysanddatamodule.attr_keys: Only relevant for computing auxiliary metrics within custom callbacks (e.g., for the Neural Latents Benchmark). Can be removed in most cases.
The following hyperparameters are also frequently changed, in order of importance:
pbt.hps: The ranges from which PBT should initialize and resample learning rates and regularization hyperparameters.trainer.max_epochs: The total number of epochs to train the model.pbt.perturbation_interval: The number of epochs between successive perturbations of the population.pbt.burn_in_period: The number of epochs to wait before the first hyperparameter perturbation.model.gen_dim: The size of the generator RNN.model.fac_dim: The size of the latent factors dimensionality bottleneck.
Advanced Configuration
The configuration system for lfads-torch is based on Hydra. Under the hood, the model, datamodule and trainer keys are recursively instantiated by passing the arguments specified to their respective _target_ objects. For example, in order for Hydra to instantiate the model, it first instantiates the readin, readout, train_aug_stack, infer_aug_stack, etc. and then passes these objects into the LFADS constructor. This is important to understand this in order to take full advantage of the modularity of lfads-torch. Augmentations, priors, and reconstruction costs can all be easily swapped out in this way to develop new functionality. See the GitHub repo for more detail.
Output Files
A successfully completed AutoLFADS run will return an autolfads.zip file containing the following:
fitlog.csv: A log of the many metrics reported during training. The most important metrics to pay attention to are the reconstruction losses,train/reconandvalid/recon. For most established model variants, this refers to the mean negative log-likelihood. Other metrics are clearly labeled in the header of the CSV. We recommend inspection oftrain/reconandvalid/reconto avoid common issues like underfitting and overfitting.lfads_output_sess0.h5: A copy of the original data file, combined with the outputs of the trained model. The denoised trials can be found attrain_output_paramsandvalid_output_params, along with other intermediate latent representations.model.ckpt: The parameters of the final trained model. When thelfads-torchsource code is released, the entire model can be loaded for post-hoc inspection and inference by passing the path to this checkpoint into theLFADS.load_from_checkpointfunction.
FAQ
How should I perform the training and validation split?
When splitting your data into training and validation sets, the main concern is ensuring that the sets follow the same distribution (equal representation of conditions, evenly distributed over the course of the recording session, etc.). A few common approaches are: (1) random sampling, (2) stratified random sampling by condition, or (3) assigning every $n^{th}$ trial to the validation set (e.g., every 5th trial for an 80/20 split).
What is the validation set used for? Is the model ever retrained on the combined data?
During an AutoLFADS training run, the validation set is used to periodically evaluate model performance (valid/recon_smth) so that the poor-performing models can be replaced with perturbed variants of the high-performing models. While we like the idea of retraining the model with the validation set included, this doesn't quite fit into the population-based training framework. However, the validation set performance will somewhat steer optimization since it is used for model selection at regular intervals.
How should I split my data into encod_data and recon_data?
The encod_data and recon_data represent the input to the model and the data against which model output is compared, respectively. In the original implementation of AutoLFADS, these were assumed to be the same. Here, we took a more flexible approach that specifies encod_data and recon_data separately to enable prediction of held-out neurons (encod_data_dim < recon_data_dim), prediction of held-out time steps (encod_seq_len < recon_seq_len), and prediction across data modalities. If you want to use the original functionality, you can should save the same data under two different keys (e.g., train_encod_data == train_recon_data).
How can I include external inputs to the generator?
External inputs to the generator can be provided using the train_ext_input and valid_ext_input keys. You'll also need to update model.ext_input_dim in the config to the appropriate input dimensionality.
How can I be sure that the model is fitting my data well?
The most important metrics to monitor are train/recon and valid/recon (the Poisson negative log-likelihood). These can be plotted from the fitlog.csv using a function like the example below.
import pandas as pd
import matplotlib.pyplot as plt
def plot_metric(log_path, metric, ylims):
df = pd.read_csv(log_path, index_col=0)
fig, axes = plt.subplots(ncols=2, figsize=(10, 5), sharex=True, sharey=True)
for id, worker in df.groupby("worker_id"):
axes[0].plot(worker.epoch, worker[f"train/{metric}"])
axes[1].plot(worker.epoch, worker[f"valid/{metric}"])
for ax in axes:
ax.set_xlabel("epoch")
ax.set_ylabel(metric)
ax.grid()
axes[0].set_title("train")
axes[1].set_title("valid")
plt.ylim(*ylims)
plt.tight_layout()
plot_metric("fitlog.csv", "recon", (0.085, 0.092))
When visualizing these curves, you are mainly looking for signs of underfitting (e.g. validation NLL is still going down and the run has stopped prematurely) or overfitting (e.g. validation NLL has started increasing and the run has gone on too long). Automatic stopping should prevent overfitting, but underfitting might require adjusting architecture or HP ranges. Below is an example of a healthy NLL curve.
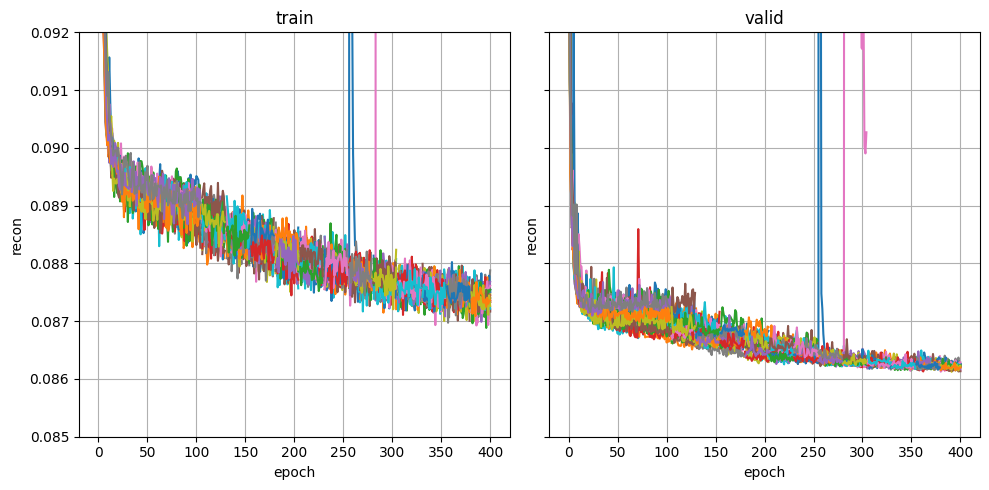
Which model is used at the end of training?
The final model is the member of the population with the lowest validation NLL.
What GPU resources are available and what are the constraints on the number of workers per GPU?
The AutoLFADS image uses p2.8xlarge instances on AWS, which have 8 GPUs each with about 12GB of memory per GPU. We typically run two models per GPU, but you may run more or less depending on your dataset and model size. It's worth noting that PBT can handle asynchronous generations so you could easily run e.g. 32 workers, keeping 2 workers per GPU but it would just take twice as long as 16. Checking GPU usage on the instance to confirm that your models will fit may be challenging, and we recommend contacting NeuroCAAS if this is critical.
Useful Links
AutoLFADS (Keshtkaran*, Sedler* et al. 2022 and Sedler et al. 2023) Paper Link
AutoLFADS (Keshtkaran*, Sedler* et al. 2022 and Sedler et al. 2023) Github Repo Link
AutoLFADS (Keshtkaran*, Sedler* et al. 2022 and Sedler et al. 2023) Bash Script Link
AutoLFADS (Keshtkaran*, Sedler* et al. 2022 and Sedler et al. 2023) Demo Link