LFADS
Latent Factor Analysis via Dynamical Systems
Abstract
Neuroscience is experiencing a revolution in which simultaneous recording of thousands of neurons is revealing population dynamics that are not apparent from single-neuron responses. This structure is typically extracted from data averaged across many trials, but deeper understanding requires studying phenomena detected in single trials, which is challenging due to incomplete sampling of the neural population, trial-to-trial variability, and fluctuations in action potential timing. We introduce latent factor analysis via dynamical systems, a deep learning method to infer latent dynamics from single-trial neural spiking data. When applied to a variety of macaque and human motor cortical datasets, latent factor analysis via dynamical systems accurately predicts observed behavioral variables, extracts precise firing rate estimates of neural dynamics on single trials, infers perturbations to those dynamics that correlate with behavioral choices, and combines data from non-overlapping recording sessions spanning months to improve inference of underlying dynamics.
Papers
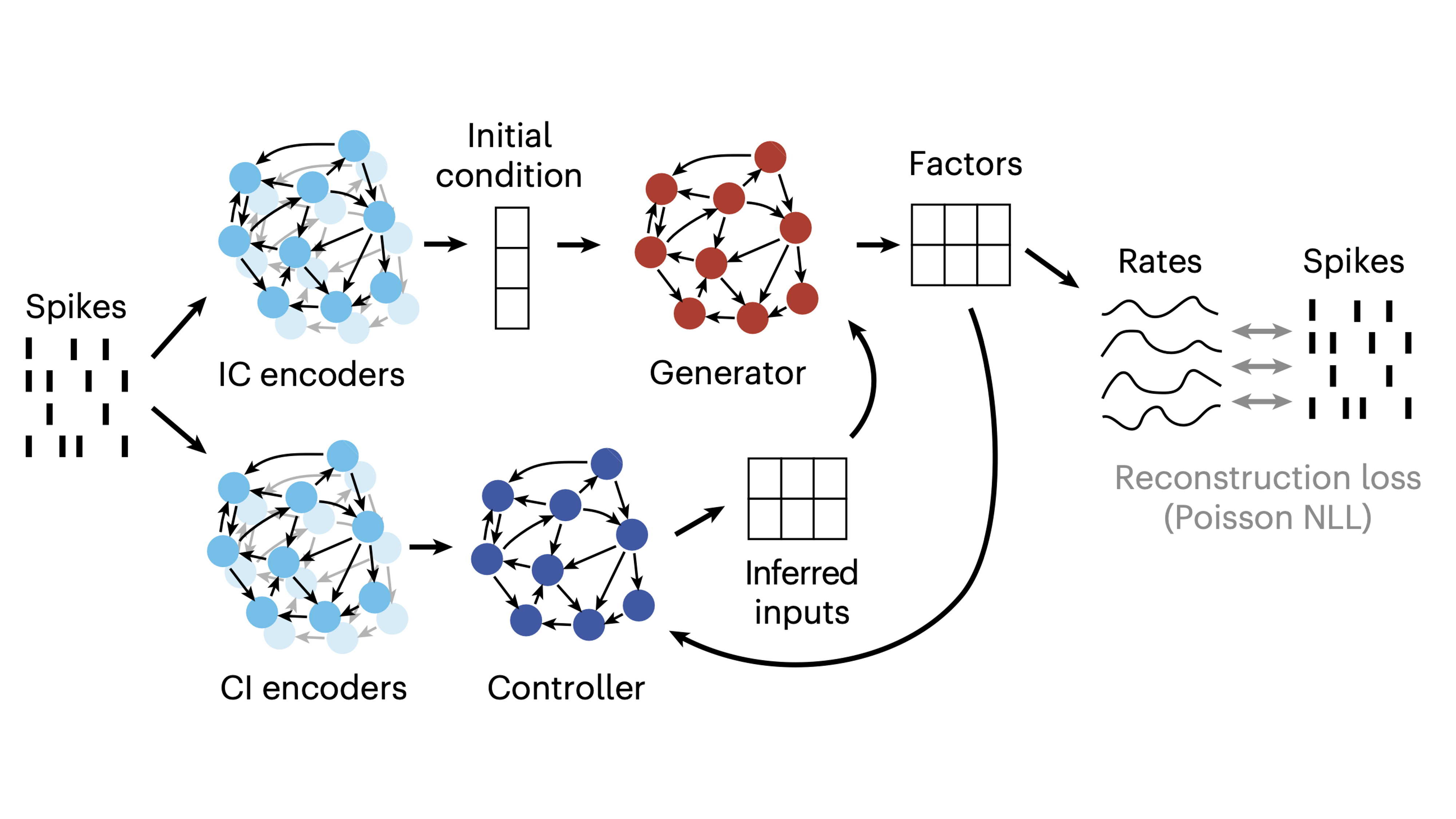
- Inferring single-trial neural population dynamics using sequential auto-encoders - Introduces LFADS with applications to neural data. Includes an in-depth techical breakdown in the appendix.
- A large-scale neural network training framework for generalized estimation of single-trial population dynamics - Presents AutoLFADS, an automatic hyperparameter tuning framework that efficiently finds good LFADS models.
- lfads-torch: A modular and extensible implementation of latent factor analysis via dynamical systems - Presents
lfads-torch, a PyTorch implementation of AutoLFADS and the flagship LFADS codebase. - High-performance neural population dynamics modeling enabled by scalable computational infrastructure - Presents methods for deploying AutoLFADS at scale.
Talks
- Methods Lecture: Deep Learning and LFADS - Given by Chethan Pandarinath at the first Data Science and AI for Neuroscience Summer School (2022) at Caltech.
- Latent variable modeling of neural population dynamics - where do we go from here? - Given by Chethan Pandarinath at Neuromatch 4.0.
- A large-scale neural network training framework for generalized estimation of single-trial population dynamics - Given by Andrew Sedler at Neuromatch 3.0.
- Inferring Single-Trial Neural Population Dynamics Using Sequential Auto-Encoders - Given by David Sussillo at Nature Methods 2018.
Tutorials
- Guide to Multisession Modeling with
lfads-torch- Topics:- Formatting data inputs to train a multisession
lfads-torchmodel via neural stitching - Training a model on multisession data
- Evaluating the resulting latent factors and firing rates inferred by
lfads-torch
- Formatting data inputs to train a multisession
lfads-torch Discussion on Gitter
lfads-torch Discussion on GitterThe public Gitter room provides an opportunity to discuss lfads-torch with the developers and follow pull requests, issues, and releases. To join the Gitter room, click the button below and sign up for an Element account via your GitHub credentials. You can also join via Matrix.
AutoLFADS on NeuroCAAS
NeuroCAAS is a cloud platform that connects users with Amazon Web Services cloud computing resources via a simple web interface. We have implemented AutoLFADS to run on NeuroCAAS with support for standard and multisession model training.
Citations
@article{Pandarinath_O’Shea_Collins_Jozefowicz_Stavisky_Kao_Trautmann_Kaufman_Ryu_Hochberg_et al._2018,
title={Inferring single-trial neural population dynamics using sequential auto-encoders},
volume={15},
DOI={10.1038/s41592-018-0109-9},
issue={10},
journal={Nature Methods},
author={Pandarinath, Chethan and O’Shea, Daniel J. and Collins, Jasmine and Jozefowicz, Rafal and Stavisky, Sergey D. and Kao, Jonathan C. and Trautmann, Eric M. and Kaufman, Matthew T. and Ryu, Stephen I. and Hochberg, Leigh R. and et al.},
year={2018},
month={Sep},
pages={805–815}}
@article{Keshtkaran_Sedler_Chowdhury_Tandon_Basrai_Nguyen_Sohn_Jazayeri_Miller_Pandarinath_2022,
title={A large-scale neural network training framework for generalized estimation of single-trial population dynamics},
volume={19},
DOI={10.1038/s41592-022-01675-0},
issue={12},
journal={Nature Methods},
author={Keshtkaran, Mohammad Reza and Sedler, Andrew R. and Chowdhury, Raeed H. and Tandon, Raghav and Basrai, Diya and Nguyen, Sarah L. and Sohn, Hansem and Jazayeri, Mehrdad and Miller, Lee E. and Pandarinath, Chethan},
year={2022},
month={Nov},
pages={1572–1577}}